MolMod User Guide
Step-by-step instructions for site-directed molecular optimization
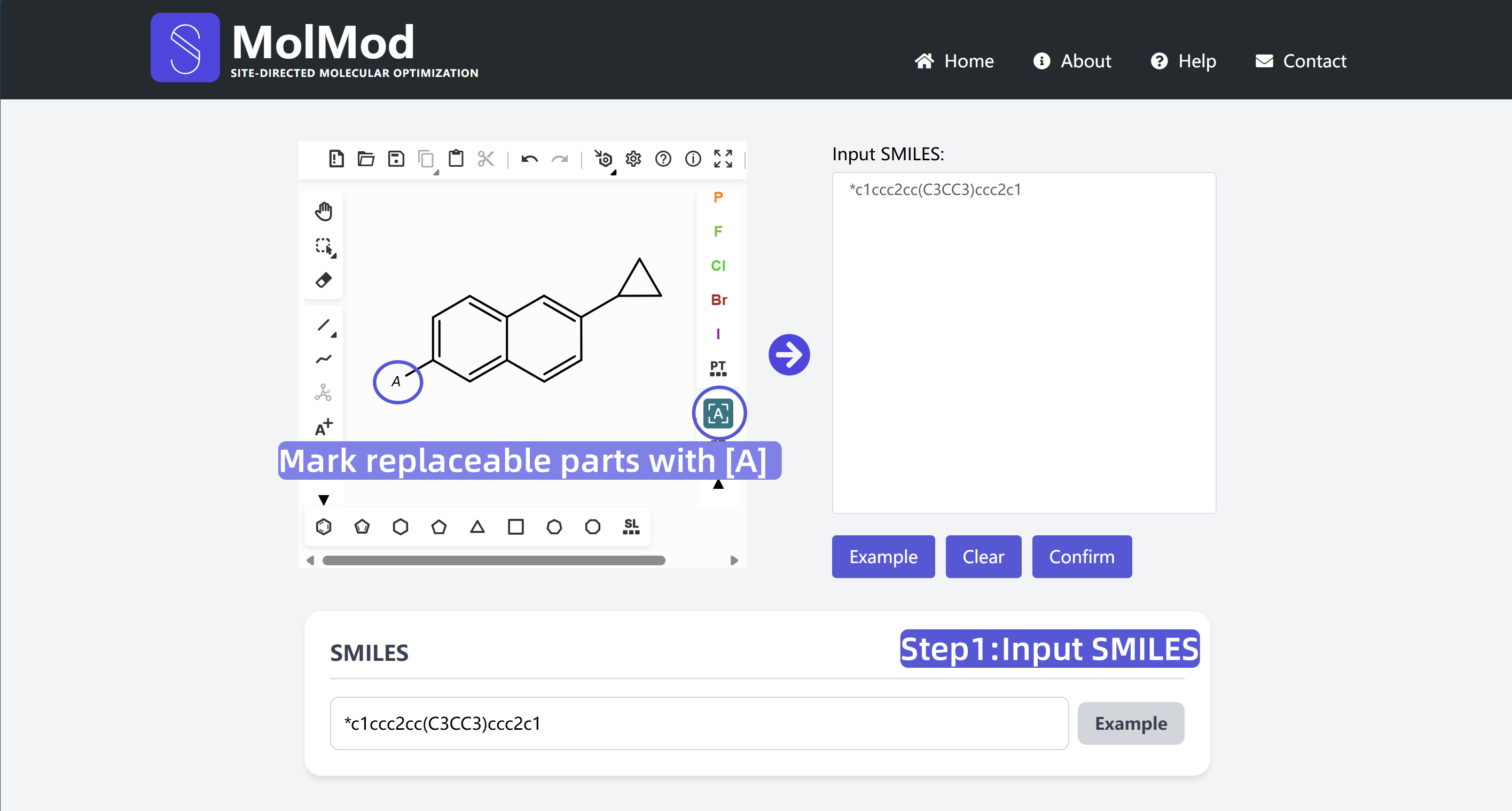
1 Input SMILES
Draw or paste molecular structure, use [A] to mark replaceable parts. The system will automatically generate the corresponding SMILES string.
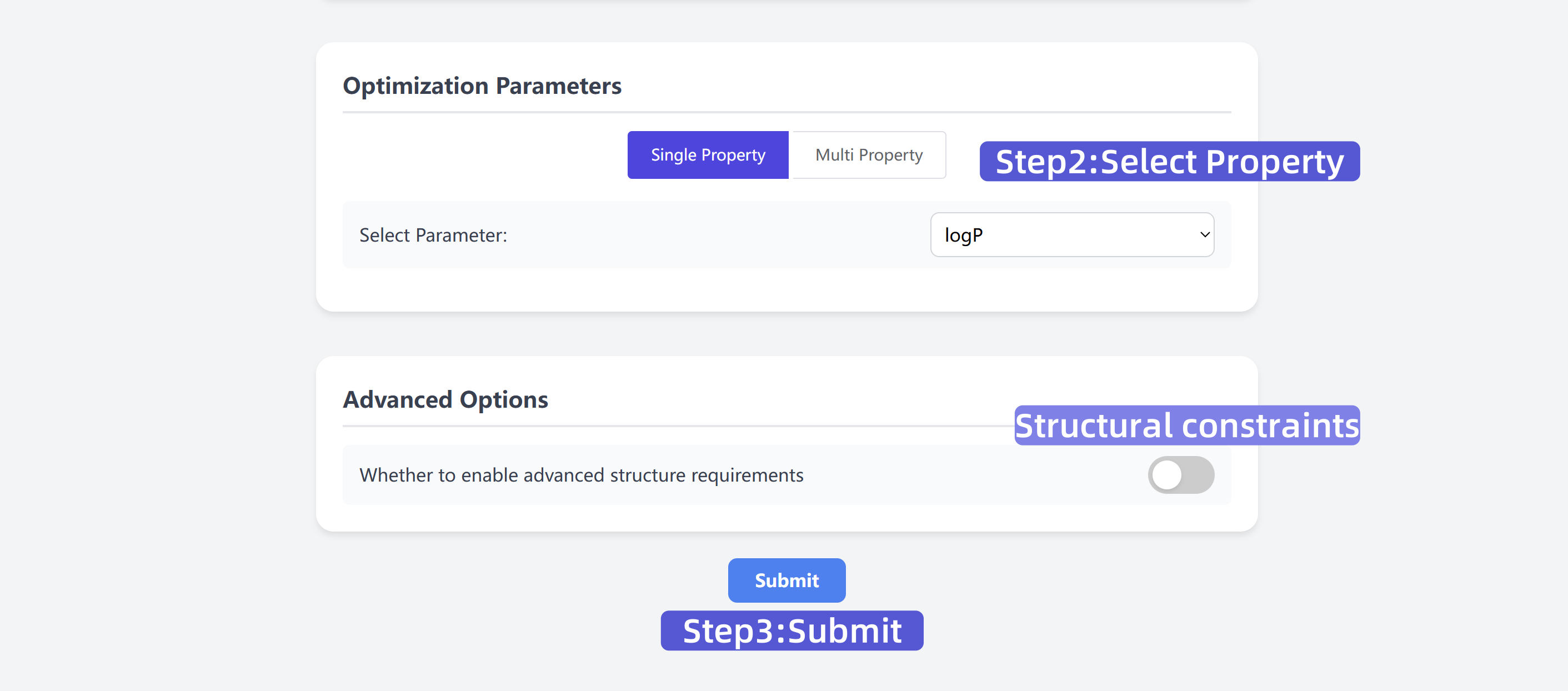
2 Select Optimization Parameters
Choose target properties to optimize, such as logP values or multi-property mode. Optionally enable structural constraints.
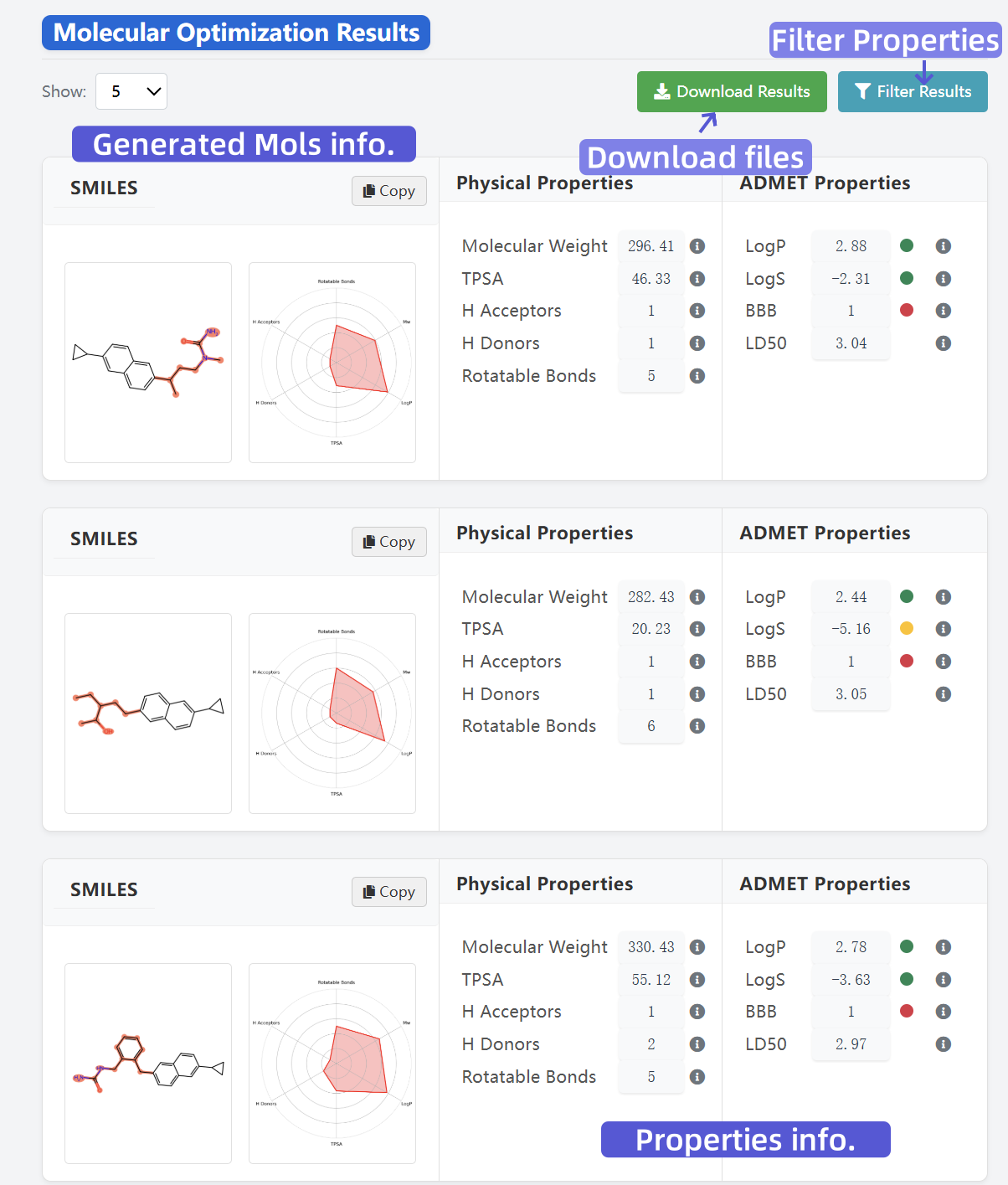
3 View and Filter Results
After submission, view generated molecules, analyze ADMET and physicochemical properties, and download result data.